Bitextor is a tool to automatically harvest bitexts from multilingual websites. To run it, it is necessary to provide:
- The source where the parallel data will be searched: one or more websites (namely, Bitextor needs website hostnames or WARC files)
- The two languages on which the user is interested: language IDs must be provided following the ISO 639-1
- A source of bilingual information between these two languages: either a bilingual lexicon (such as those available at the bitextor-data repository), a machine translation (MT) system, or a parallel corpus to be used to produce either a lexicon or an MT system (depending on the alignment strategy chosen, see below)
Installation
Bitextor can be installed via Docker, Conda or built from source. See instructions here.
Usage
usage: bitextor [-C FILE [FILE ...]] [-c KEY=VALUE [KEY=VALUE ...]]
[-j JOBS] [-k] [--notemp] [--dry-run]
[--forceall] [--forcerun [TARGET [TARGET ...]]]
[-q] [-h]
launch Bitextor
Bitextor config::
-C FILE [FILE ...], --configfile FILE [FILE ...]
Bitextor YAML configuration file
-c KEY=VALUE [KEY=VALUE ...], --config KEY=VALUE [KEY=VALUE ...]
Set or overwrite values for Bitextor config
Optional arguments::
-j JOBS, --jobs JOBS Number of provided cores
-k, --keep-going Go on with independent jobs if a job fails
--notemp Disable deletion of intermediate files marked as temporary
--dry-run Do not execute anything and display what would be done
--forceall Force rerun every job
--forcerun TARGET [TARGET ...]
List of files and rules that shall be re-created/re-executed
-q, --quiet Do not print job information
-h, --help Show this help message and exit
Advanced usage
Bitextor uses Snakemake to define Bitextor's workflow and manage its execution. Snakemake provides a lot of flexibility in terms of configuring the execution of the pipeline. For advanced users that want to make the most out of this tool, bitextor-full command is provided that calls Snakemake CLI with Bitextor's workflow and exposes all of Snakemake's parameters.
Execution on a cluster
To run Bitextor on a cluster with a software that allows to manage job queues, it is recommended to use bitextor-full command and use Snakemake's cluster configuration.
Bitextor configuration
Bitextor uses a configuration file to define the variables required by the pipeline. Depending on the options defined in this configuration file the pipeline can behave differently, running alternative tools and functionalities. For more information consult this exhaustive overview of all the options that can be set in the configuration file and how they affect the pipeline.
Suggestion: A configuration wizard called bitextor-config gets installed with Bitextor to help with this task. Furthermore, a minimalist configuration file sample is provided in this repository. You can take it as an starting point by changing all the paths to match your environment.
Bitextor output
Bitextor generates the final parallel corpora in multiple formats. These files will be placed in permanentDir folder and will have the following format: {lang1}-{lang2}.{prefix}.gz, where {prefix} corresponds to a descriptor of the corresponding format. The list of files that may be produced is the following:
{lang1}-{lang2}.raw.gz- default (always generated){lang1}-{lang2}.sent.gz- default{lang1}-{lang2}.not-deduped.tmx.gz- generated iftmx: true{lang1}-{lang2}.deduped.tmx.gz- generated ifdeduped: true{lang1}-{lang2}.deduped.txt.gz- generated ifdeduped: true{lang1}-{lang2}.not-deduped.roamed.tmx.gz- generated ifbiroamer: trueandtmx: true{lang1}-{lang2}.deduped.roamed.tmx.gz- generated ifbiroamer: trueanddeduped: true
See detailed description of the output files.
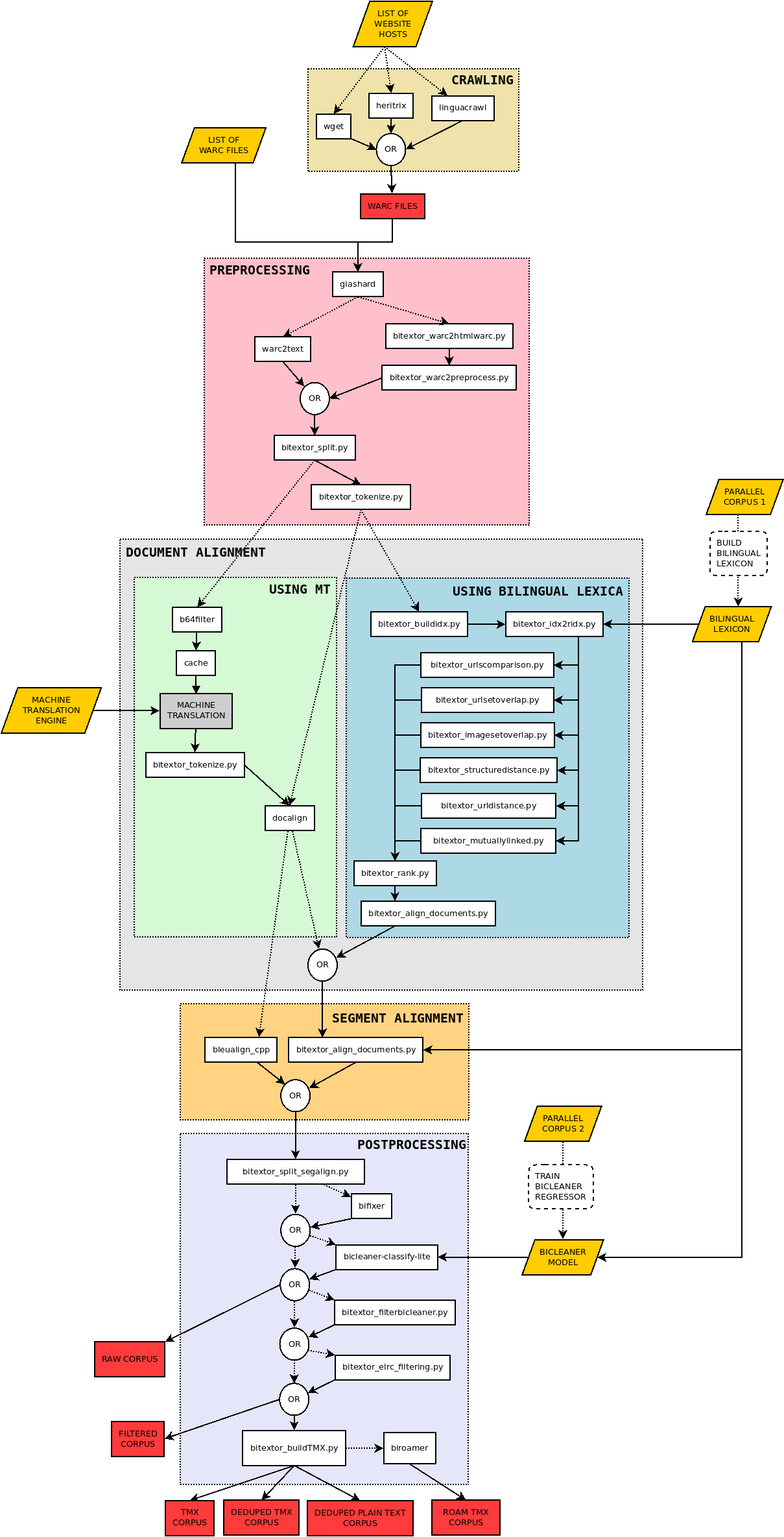
Pipeline description
Bitextor is a pipeline that runs a collection of scripts to produce a parallel corpus from a collection of multilingual websites. The pipeline is divided in five stages:
- Crawling: documents are downloaded from the specified websites
- Pre-processing: downloaded documents are normalized, boilerplates are removed, plain text is extracted, and language is identified
- Document alignment: parallel documents are identified. Two strategies are implemented for this stage:
- one using bilingual lexica and a collection of features extracted from HTML; a linear regressor combines these resources to produce a score in [0,1], and
- another using machine translation and a TF/IDF strategy to score document pairs
- Segment alignment: each pair of documents is processed to identify parallel segments. Again, two strategies are implemented:
- Post-processing: final steps that allow to clean the parallel corpus obtained using the tool Bicleaner, deduplicates translation units, and computes additional quality metrics
The following diagram shows the structure of the pipeline and the different scripts that are used in each stage:

All documents and software contained in this repository reflect only the authors' view. The Innovation and Networks Executive Agency of the European Union is not responsible for any use that may be made of the information it contains.


